Chapman - Figure 9 - HDL subspeciation Text
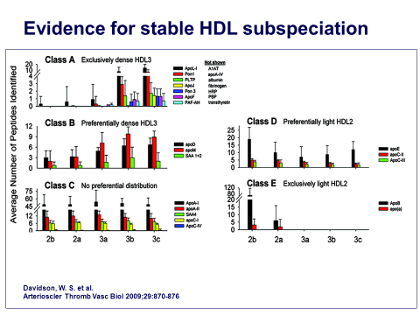
Now, having established that all of the 5 different sizes of HDL particles, from HDL2b down to HDL3c, are basically composed of 4 identical apoA-I units, the next question was which other proteins, in addition to apoA-I and apoA-II, were associated with this spectrum of HDL particle sizes. So, in collaboration with Sean Davidson’s group, my group performed an analysis of all these proteins, ie, “proteomic analyses” by mass spectrometry, and basically determined the sequences of the different peptides obtained using proteolysis with trypsin. This enabled us to determine the sequence of all those peptides and to fit them together in such a way as to tell which proteins were present in which different subpopulations of HDL particles.
As shown in the Figure, the result was 5 distinct patterns, which we have called class A, B, C, D, and E. However, the pattern that interested us the most was pattern A, because this pattern was found exclusively on the smallest, dense HDL3 particles, which are the ones that are especially active in effluxing cholesterol and in exerting anti-inflammatory and antioxidative activities.
The other reason was that many of the small proteins appear only in low abundance in plasma; for example, apo-A5, apo-F, apo-L1 are all present at 1000 to 10,000 times lower concentrations than the most abundant protein in plasma, albumin. In fact, sometimes their concentration is so low that they can only be present on a minority of HDL particles, so they cannot be very contributory to the major functions of HDL. So the next question was, is there a pattern of associations among these proteins in the Class A pattern?
J Clin Lipidol. 2011; 5(6).